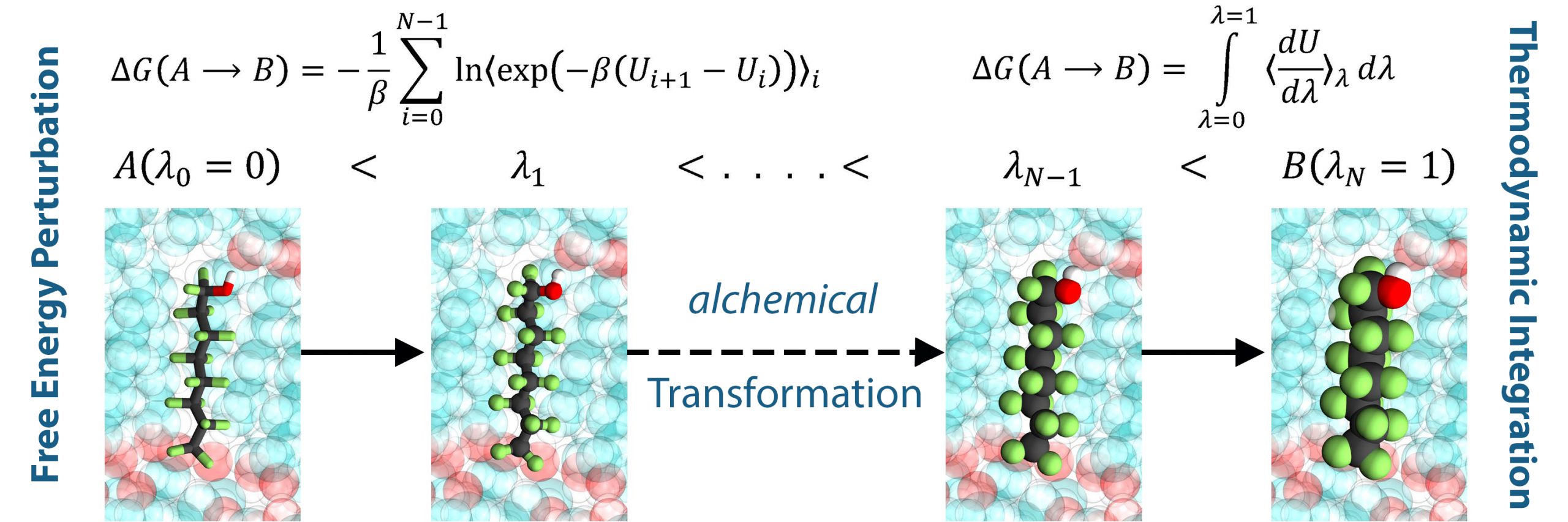
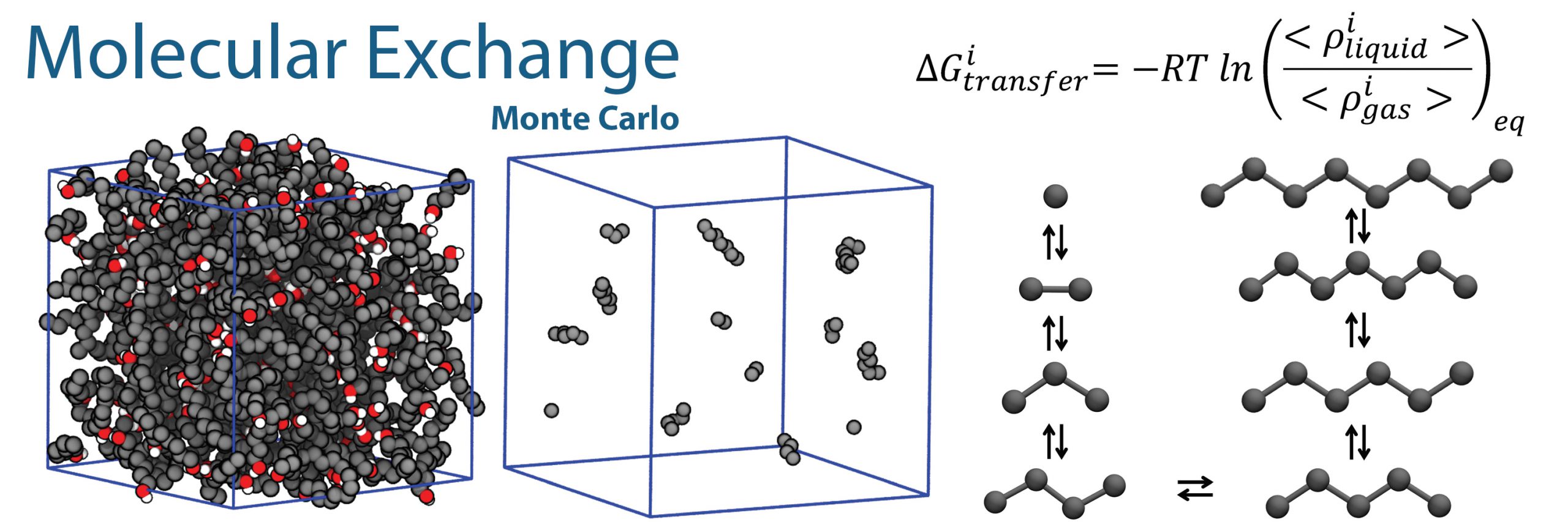
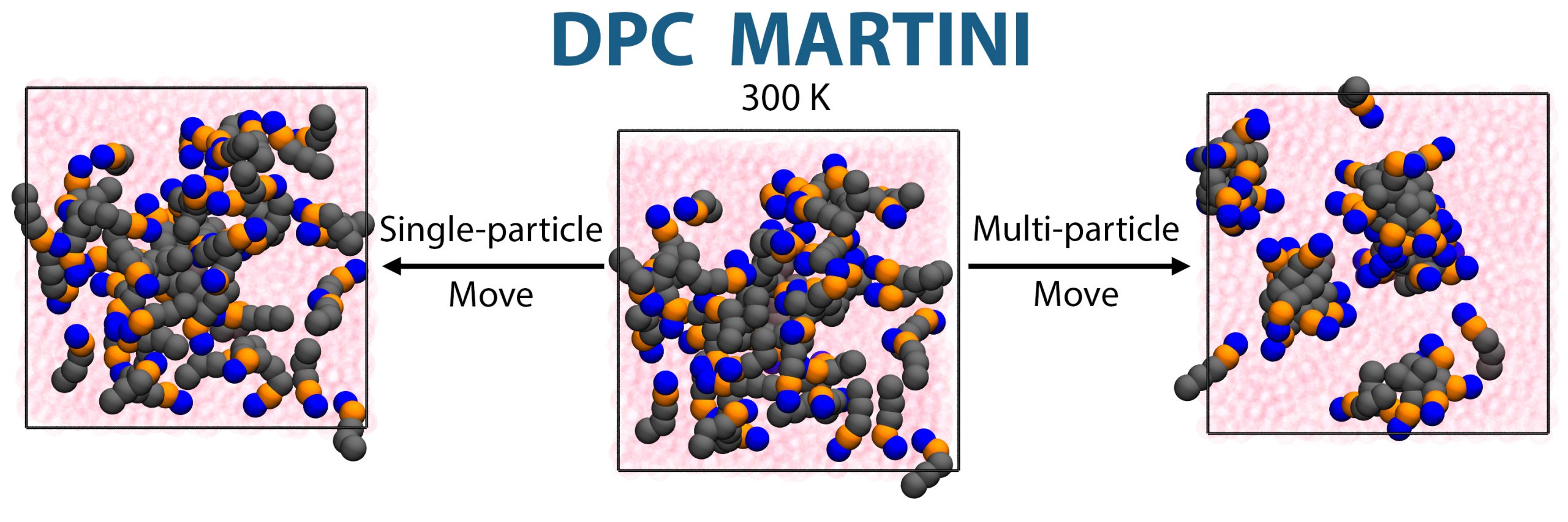
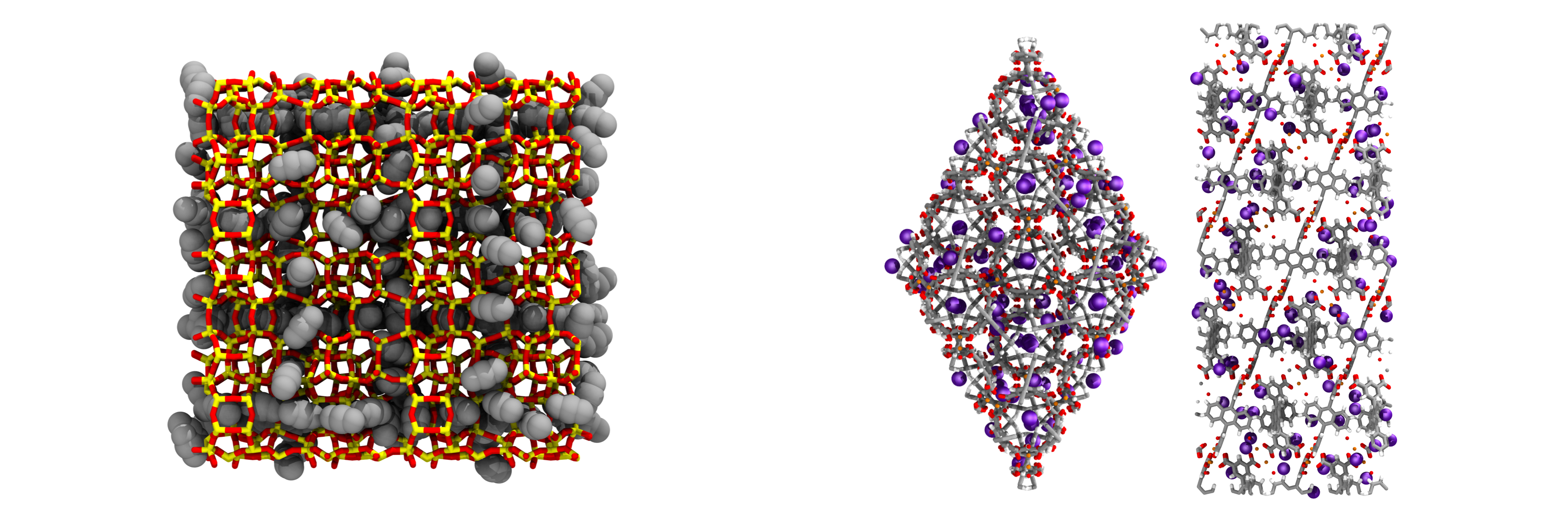
GPU Optimized Monte Carlo (GOMC)
GOMC is open-source software for simulating molecular systems using the Metropolis Monte Carlo algorithm. The software has been written in object-oriented C++, and uses OpenMP and NVIDIA CUDA to allow for execution on multicore CPU and GPU architectures, respectively. GOMC employs widely-used simulation file types (PDB, PSF, CHARMM-style parameter files). GOMC can be used to study vapor–liquid equilibria, adsorption in porous materials, surfactant self-assembly, and condensed phase structure for complex molecules. To learn more about GOMC, please refer to our documentation and our most recently published description of the GOMC software.
Ensembles
GOMC supports simulations in a variety of ensembles, which include:
Force fields
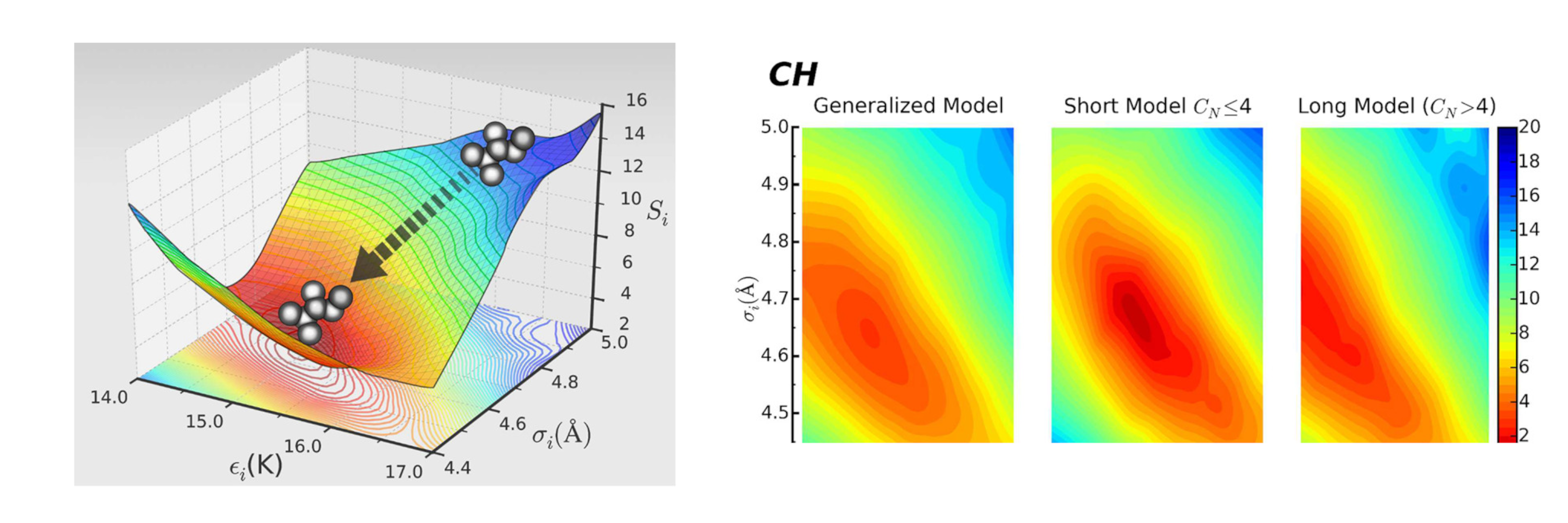
GOMC supports a variety of all-atom, united atom, and coarse grained force fields such as:
Molecule
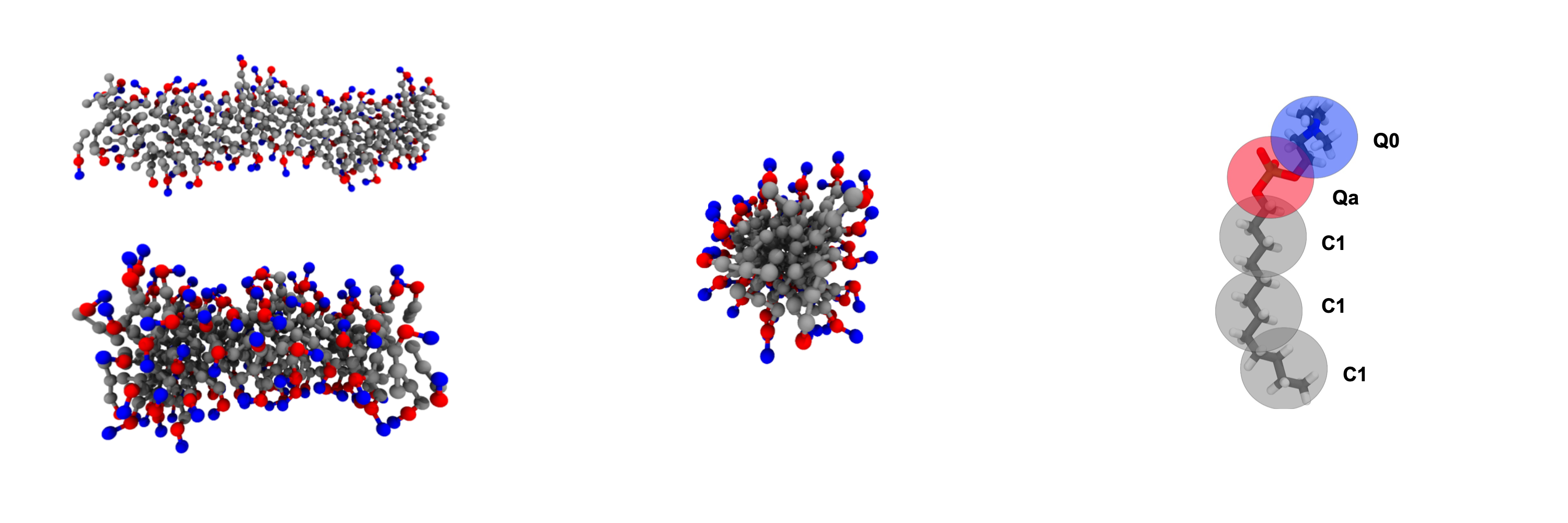
GOMC supports a variety of molecular topologies:
Monte Carlo moves
GOMC supports a variety of Monte Carlo moves, such as: